SAFlex.Beta
The SAFlex.Beta structural letters
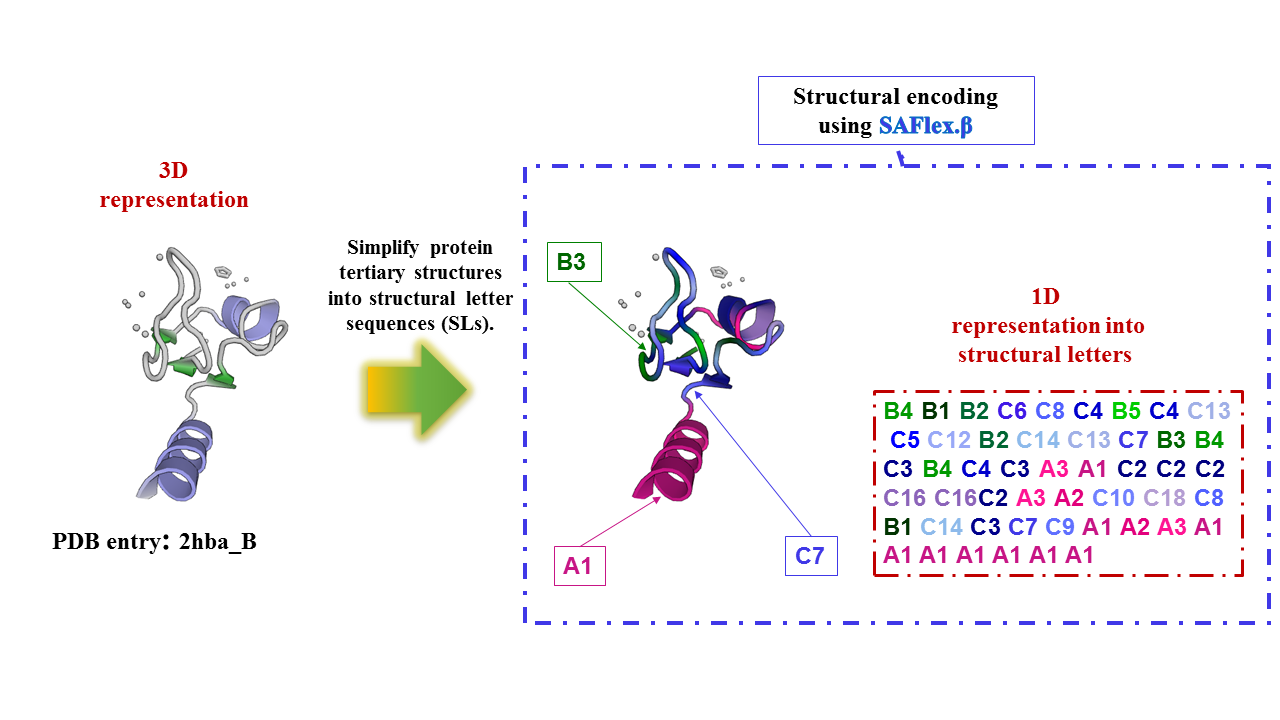
A structural letter (SL) is a representative protein fragment, namely it is a recurrent structural 3D building block. It consists of four amino acid residues in a protein. Each protein fragment is described by a vector of four descriptors (d1,d2,d3 and d4 ), that provide a unique representation of the 3D structure of the protein fragment. The 27 identified SA-letters are symbolized by A1,A2,..., B1,B2,..., C1,...,C19. The collection of SA-letters compose the structural alphabet SAFlex.Beta.
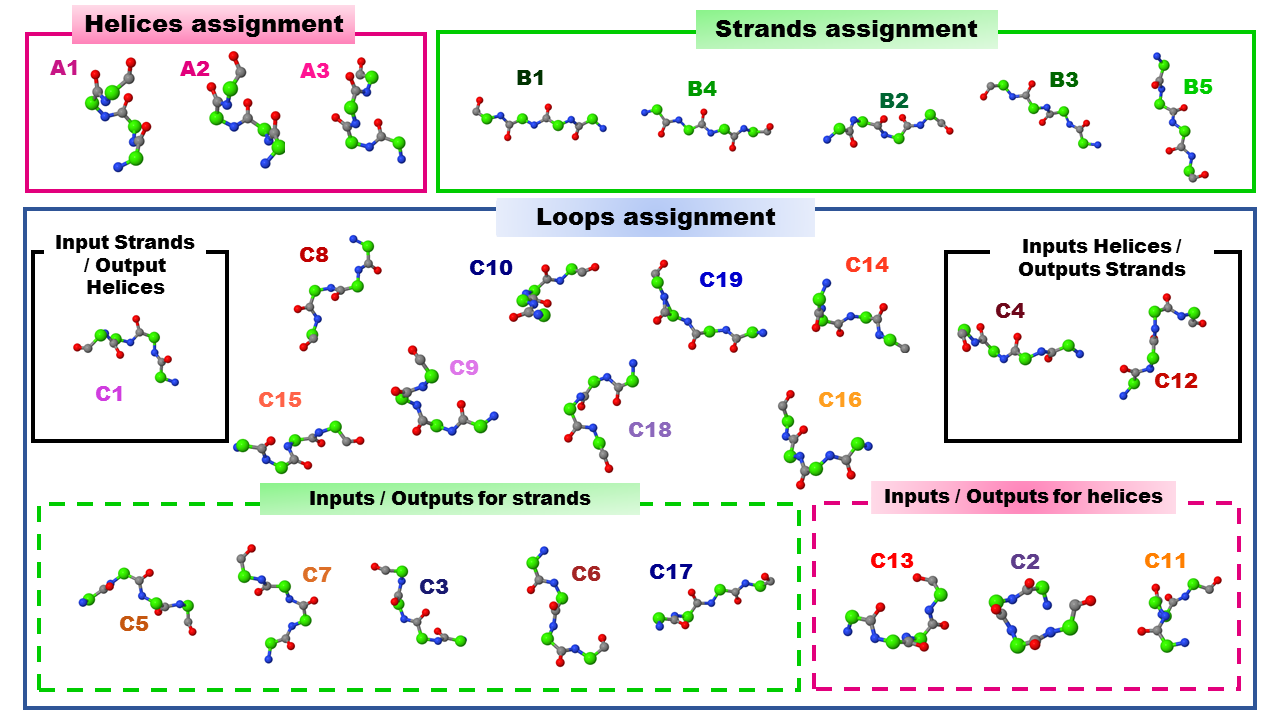
Structural letter parameters
| Structural letter | Frequency(%) |
|---|---|
| 16.9927 | |
| 12.0367 | |
| 5.3991 | |
| 6.3639 | |
| 5.0177 | |
| 4.7882 | |
| 4.6072 | |
| 4.5312 | |
| 3.9468 | |
| 3.3261 | |
| 2.9728 | |
| 2.9488 | |
| 2.9419 | |
| 2.9016 | |
| 2.6179 | |
| 2.4214 | |
| 2.2669 | |
| 2.1289 | |
| 1.9340 | |
| 1.8567 | |
| 1.7619 | |
| 1.5074 | |
| 1.4943 | |
| 1.1900 | |
| 1.0912 | |
| 0.6158 | |
| 0.3389 |
Structural letter A1
$$ μ_{A1}=
\begin{pmatrix}
5.45 & 5.12 & 5.45 & 2.93 \\
\end{pmatrix}
$$
$$ Σ_{A1}=
\begin{pmatrix}
0.011 & 0.007 & -0.001 & -0.003 \\
0.007 & 0.025 & 0.004 & 0.014 \\
-0.001 & 0.004 & 0.01 & -0.005 \\
-0.003 & 0.014 & -0.005 & 0.026 \\
\end{pmatrix}
$$
$$ μ_{A2}=
\begin{pmatrix}
5.5 & 5.25 & 5.52 & 2.85 \\
\end{pmatrix}
$$
$$ Σ_{A2}=
\begin{pmatrix}
0.033 & 0.021 & -0.005 & -0.009 \\
0.021 & 0.065 & 0.016 & 0.032 \\
-0.005 & 0.016 & 0.033 & -0.016 \\
-0.009 & 0.032 & -0.016 & 0.073 \\
\end{pmatrix}
$$
$$ μ_{A3}=
\begin{pmatrix}
5.42 & 5.63 & 5.42 & 3.41 \\
\end{pmatrix}
$$
$$ Σ_{A3}=
\begin{pmatrix}
0.035 & 0.024 & -0.002 & -0.005 \\
0.024 & 0.078 & 0.01 & 0.032 \\
-0.002 & 0.01 & 0.027 & -0.009 \\
-0.005 & 0.032 & -0.009 & 0.032 \\
\end{pmatrix}
$$
$$ μ_{B1}=
\begin{pmatrix}
6.81 & 10.23 & 6.69 & -0.71 \\
\end{pmatrix}
$$
$$ Σ_{B1}=
\begin{pmatrix}
0.065 & 0.067 & 0.039 & -0.002 \\
0.067 & 0.1 & 0.079 & 0.037 \\
0.039 & 0.079 & 0.075 & 0.031 \\
-0.002 & 0.037 & 0.031 & 0.264 \\
\end{pmatrix}
$$
$$ μ_{B2}=
\begin{pmatrix}
6.62 & 9.34 & 6.44 & -2.98 \\
\end{pmatrix}
$$
$$ Σ_{B2}=
\begin{pmatrix}
0.095 & 0.063 & 0.006 & -0.029 \\
0.063 & 0.115 & 0.055 & 0.057 \\
0.006 & 0.055 & 0.055 & 0.041 \\
-0.029 & 0.057 & 0.041 & 0.102 \\
\end{pmatrix}
$$
$$ μ_{B3}=
\begin{pmatrix}
6.32 & 9.81 & 6.71 & -1.27 \\
\end{pmatrix}
$$
$$ Σ_{B3}=
\begin{pmatrix}
0.118 & 0.111 & 0.05 & -0.057 \\
0.111 & 0.129 & 0.074 & 0.009 \\
0.05 & 0.074 & 0.07 & 0.001 \\
-0.057 & 0.009 & 0.001 & 0.334 \\
\end{pmatrix}
$$
$$ μ_{B4}=
\begin{pmatrix}
6.88 & 9.88 & 6.47 & -2.21 \\
\end{pmatrix}
$$
$$ Σ_{B4}=
\begin{pmatrix}
0.069 & 0.064 & 0.041 & -0.03 \\
0.064 & 0.118 & 0.092 & 0.041 \\
0.041 & 0.092 & 0.085 & 0.014 \\
-0.03 & 0.041 & 0.014 & 0.182 \\
\end{pmatrix}
$$
$$ μ_{B5}=
\begin{pmatrix}
6.54 & 10.23 & 7.03 & 0.33 \\
\end{pmatrix}
$$
$$ Σ_{B5}=
\begin{pmatrix}
0.082 & 0.075 & 0.015 & 0.03 \\
0.075 & 0.074 & 0.022 & 0.017 \\
0.015 & 0.022 & 0.028 & 0.021 \\
0.03 & 0.017 & 0.021 & 0.28 \\
\end{pmatrix}
$$
$$ μ_{C1}=
\begin{pmatrix}
5.63 & 8.2 & 6.65 & 3.01 \\
\end{pmatrix}
$$
$$ Σ_{C1}=
\begin{pmatrix}
0.06 & 0.063 & 0.001 & 0.003 \\
0.063 & 0.213 & 0.043 & -0.084 \\
0.001 & 0.043 & 0.087 & -0.088 \\
0.003 & -0.084 & -0.088 & 0.129 \\
\end{pmatrix}
$$
$$ μ_{C2}=
\begin{pmatrix}
5.65 & 5.47 & 5.94 & 1.94 \\
\end{pmatrix}
$$
$$ Σ_{C2}=
\begin{pmatrix}
0.068 & 0.079 & -0.003 & -0.026 \\
0.079 & 0.191 & 0.056 & -0.016 \\
-0.003 & 0.056 & 0.054 & -0.05 \\
-0.026 & -0.016 & -0.05 & 0.249 \\
\end{pmatrix}
$$
$$ μ_{C3}=
\begin{pmatrix}
6.78 & 8.91 & 6.4 & -3.38 \\
\end{pmatrix}
$$
$$ Σ_{C3}=
\begin{pmatrix}
0.099 & 0.095 & -0 & 0.002 \\
0.095 & 0.207 & 0.062 & 0.048 \\
-0 & 0.062 & 0.054 & 0.043 \\
0.002 & 0.048 & 0.043 & 0.044 \\
\end{pmatrix}
$$
$$ μ_{C4}=
\begin{pmatrix}
6.63 & 8.68 & 5.53 & -3 \\
\end{pmatrix}
$$
$$ Σ_{C4}=
\begin{pmatrix}
0.124 & 0.057 & 0.014 & -0.044 \\
0.057 & 0.099 & 0.057 & 0.029 \\
0.014 & 0.057 & 0.06 & -0.012 \\
-0.044 & 0.029 & -0.012 & 0.135 \\
\end{pmatrix}
$$
$$ μ_{C5}=
\begin{pmatrix}
5.67 & 9.07 & 6.6 & 1.78 \\
\end{pmatrix}
$$
$$ Σ_{C5}=
\begin{pmatrix}
0.057 & 0.048 & 0.008 & 0.016 \\
0.048 & 0.082 & 0.031 & -0.108 \\
0.008 & 0.031 & 0.079 & -0.014 \\
0.016 & -0.108 & -0.014 & 0.486 \\
\end{pmatrix}
$$
$$ μ_{C6}=
\begin{pmatrix}
6.52 & 9.16 & 5.62 & -1.3 \\
\end{pmatrix}
$$
$$ Σ_{C6}=
\begin{pmatrix}
0.124 & 0.064 & 0.024 & -0.028 \\
0.064 & 0.095 & 0.07 & 0.056 \\
0.024 & 0.07 & 0.068 & 0.015 \\
-0.028 & 0.056 & 0.015 & 0.418 \\
\end{pmatrix}
$$
$$ μ_{C7}=
\begin{pmatrix}
5.72 & 7.34 & 7.02 & 1.04 \\
\end{pmatrix}
$$
$$ Σ_{C7}=
\begin{pmatrix}
0.083 & 0.137 & 0.009 & 0 \\
0.137 & 0.386 & 0.074 & 0.022 \\
0.009 & 0.074 & 0.041 & -0.111 \\
0 & 0.022 & -0.111 & 1.138 \\
\end{pmatrix}
$$
$$ μ_{C8}=
\begin{pmatrix}
6.1 & 9.16 & 5.79 & 0.63 \\
\end{pmatrix}
$$
$$ Σ_{C8}=
\begin{pmatrix}
0.17 & 0.135 & 0.056 & 0.053 \\
0.135 & 0.153 & 0.102 & 0.011 \\
0.056 & 0.102 & 0.099 & 0.03 \\
0.053 & 0.011 & 0.03 & 0.551 \\
\end{pmatrix}
$$
$$ μ_{C9}=
\begin{pmatrix}
6.45 & 5.9 & 5.57 & 0.75 \\
\end{pmatrix}
$$
$$ Σ_{C9}=
\begin{pmatrix}
0.09 & 0.157 & -0.004 & 0.041 \\
0.157 & 0.389 & 0.048 & 0.248 \\
-0.004 & 0.048 & 0.035 & 0.042 \\
0.041 & 0.248 & 0.042 & 1.097 \\
\end{pmatrix}
$$
$$ μ_{C10}=
\begin{pmatrix}
6.94 & 8.11 & 6.22 & -2.16 \\
\end{pmatrix}
$$
$$ Σ_{C10}=
\begin{pmatrix}
0.086 & 0.202 & 0.02 & -0.082 \\
0.202 & 1.189 & 0.468 & -0.26 \\
0.02 & 0.468 & 0.295 & 0.12 \\
-0.082 & -0.26 & 0.12 & 1.136 \\
\end{pmatrix}
$$
$$ μ_{C11}=
\begin{pmatrix}
5.58 & 7.84 & 5.62 & -3.22 \\
\end{pmatrix}
$$
$$ Σ_{C11}=
\begin{pmatrix}
0.064 & 0.048 & -0.008 & 0.003 \\
0.048 & 0.16 & -0.007 & 0.152 \\
-0.008 & -0.007 & 0.055 & -0.033 \\
0.003 & 0.152 & -0.033 & 0.226 \\
\end{pmatrix}
$$
$$ μ_{C12}=
\begin{pmatrix}
6.74 & 8.08 & 5.48 & -3.7 \\
\end{pmatrix}
$$
$$ Σ_{C12}=
\begin{pmatrix}
0.076 & 0.027 & -0.004 & -0.011 \\
0.027 & 0.083 & 0.033 & 0.008 \\
-0.004 & 0.033 & 0.041 & -0.003 \\
-0.011 & 0.008 & -0.003 & 0.011 \\
\end{pmatrix}
$$
$$ μ_{C13}=
\begin{pmatrix}
5.61 & 6.93 & 5.59 & 3.74 \\
\end{pmatrix}
$$
$$ Σ_{C13}=
\begin{pmatrix}
0.082 & 0.121 & 0.032 & -0.001 \\
0.121 & 0.357 & 0.127 & -0.009 \\
0.032 & 0.127 & 0.093 & -0.007 \\
-0.001 & -0.009 & -0.007 & 0.004 \\
\end{pmatrix}
$$
$$ μ_{C14}=
\begin{pmatrix}
5.84 & 6.48 & 6 & 2.97 \\
\end{pmatrix}
$$
$$ Σ_{C14}=
\begin{pmatrix}
0.204 & 0.205 & -0.05 & 0.024 \\
0.205 & 0.59 & 0.139 & 0.13 \\
-0.05 & 0.139 & 0.148 & -0.023 \\
0.024 & 0.13 & -0.023 & 0.167 \\
\end{pmatrix}
$$
$$ μ_{C15}=
\begin{pmatrix}
5.78 & 8.18 & 5.68 & 3.08 \\
\end{pmatrix}
$$
$$ Σ_{C15}=
\begin{pmatrix}
0.135 & 0.082 & 0.016 & 0.035 \\
0.082 & 0.171 & 0.071 & -0.096 \\
0.016 & 0.071 & 0.084 & -0.003 \\
0.035 & -0.096 & -0.003 & 0.188 \\
\end{pmatrix}
$$
$$ μ_{C16}=
\begin{pmatrix}
5.73 & 7.56 & 6.75 & -1.64 \\
\end{pmatrix}
$$
$$ Σ_{C16}=
\begin{pmatrix}
0.097 & 0.099 & -0.003 & -0.04 \\
0.099 & 1.715 & 0.376 & -0.689 \\
-0.003 & 0.376 & 0.178 & -0.021 \\
-0.04 & -0.689 & -0.021 & 1.216 \\
\end{pmatrix}
$$
$$ μ_{C17}=
\begin{pmatrix}
6.74 & 9.44 & 7.03 & 1.75 \\
\end{pmatrix}
$$
$$ Σ_{C17}=
\begin{pmatrix}
0.123 & 0.052 & -0.031 & -0.011 \\
0.052 & 0.6 & 0.145 & 0.146 \\
-0.031 & 0.145 & 0.07 & 0.021 \\
-0.011 & 0.146 & 0.021 & 0.423 \\
\end{pmatrix}
$$
$$ μ_{C18}=
\begin{pmatrix}
5.59 & 5.81 & 5.54 & -3.18 \\
\end{pmatrix}
$$
$$ Σ_{C18}=
\begin{pmatrix}
0.119 & 0.137 & 0.001 & 0.005 \\
0.137 & 0.417 & 0.032 & -0.171 \\
0.001 & 0.032 & 0.05 & 0.02 \\
0.005 & -0.171 & 0.02 & 0.179 \\
\end{pmatrix}
$$
$$ μ_{C19}=
\begin{pmatrix}
7.59 & 10.31 & 7.5 & -0.3 \\
\end{pmatrix}
$$
$$ Σ_{C19}=
\begin{pmatrix}
0.242 & 0.232 & -0.157 & -0.878 \\
0.232 & 0.624 & 0.396 & -0.412 \\
-0.157 & 0.396 & 0.42 & -0.415 \\
-0.878 & -0.412 & -0.415 & 0.508 \\
\end{pmatrix}
$$
Legend
The mean vector \(μ \in \mathbb{R}^4\) and the covariance matrix \(Σ \in \mathbb{R}^{4\times4}\) for the structural letter.
Transition matrix
Download transition matrix here
How to cite SAFlex in Publications ?
To cite SAFlex, please refer to the following publication :
